Gödel, Escher, Bach: An Eternal Golden Braid (91 page)
Read Gödel, Escher, Bach: An Eternal Golden Braid Online
Authors: Douglas R. Hofstadter
Tags: #Computers, #Art, #Classical, #Symmetry, #Bach; Johann Sebastian, #Individual Artists, #Science, #Science & Technology, #Philosophy, #General, #Metamathematics, #Intelligence (AI) & Semantics, #G'odel; Kurt, #Music, #Logic, #Biography & Autobiography, #Mathematics, #Genres & Styles, #Artificial Intelligence, #Escher; M. C

given, but there is no complementary base where the enzyme is bound at that instant, then the enzyme just detaches itself from the strand, and its job is done.
It should be mentioned that when a "cut" instruction is encountered, this pertains to both strands (if there are two): however, "delete" pertains only to the strand on which the enzyme is working. If Copy mode is on, then the "insert" command pertains to both strands-the base itself into the strand the enzyme is working on, and its complement into the other strand.
If Copy mode is
off
, then the "insert" command pertains only to the one strand, so a blank space must he inserted into the complementary strand.
And, whenever Copy mode is on, "move" and "search" commands require that one manufacture complementary bases to all bases which the sliding enzyme touches.
Incidentally, Copy mode is always
off
when an enzyme starts to work. If Copy mode is
off
, and the command "Shut off copy mode" is encountered, nothing happens. Likewise, If Copy mode is already
on
, and the command "Turn copy mode on" is encountered, then nothing happens.
Amino Acids
There are fifteen types of command, listed below:
Cut
cut strand(s)
del
delete a base from strand
swi
switch enzyme to other strand
mvr
move one unit to the right
mvl
move one unit to the left
cop
turn on Copy mode
off
turn off Copy mode
ina
insert A to the right of this unit
inc
insert C to the right of this unit
ing
insert G to the right of this unit
int
insert T to the right of this unit
rpy
search for the nearest pyrimidine to the right
rpu
search for the nearest purine to the right
Ipy
search for the nearest pyrimidine to the left
lpu
search for the nearest purine to the left
Each one has a three-letter abbreviation. We shall refer to the three-letter abbreviations of commands as amino acids. Thus,
every enzyme is made up of a sequence of amino acids
. Let us write down an arbitrary enzyme:
rpu - inc - cop - myr - tnyl - swi - Ipu - int
and an arbitrary strand:
TAGATCCAGTCCATCGA
and see how the enzyme acts on the strand. It so happens that the enzyme binds to
G
only.
Let us bind to the middle
G
and begin. Search rightwards for a purine (viz.,
A
or
G
). We (the enzyme) skip over
TCC
and land on
A.
Insert a
C
. Now we have
TAGATCCAGTCCACTCGA
where the arrow points to the unit to which the enzyme is bound. Set Copy mode. This puts an upside-down
G
above the
C
. Move right, move left, then switch to the other strand. Here's what we have so far:
7V
TAGATCCAGTCCACTCGA
Let's turn it upside down, 'so that the enzyme is attached to the lower strand:
VDJIDV»1DVDD1VDV1
AG
Now we search leftwards for a purine and find A. Copy mode is on, but the complementary bases are already there, so nothing is added. Finally, we insert a T (in Copy mode), and quit:
VD)IVJVJJl7V)DIVDV1
ATG
Our final product is thus two strands:
ATG
, and
TAGATCCAGTCCACATCGA
The old one is of course gone.
Translation and the Typogenetic Code
Now you might be wondering where the enzymes and strands come from, and how to tell the initial binding-preference of a given enzyme. One way might be just to throw some random strands and some random enzymes together, and see what happens when those enzymes act on those strands and their progeny. This has a similar flavor to the MU-puzzle, where there were some given rules of inference and an axiom, and you just began. The only difference is that here, every time a strand is acted on, its original form is gone forever. In the
MU
-puzzle, acting on
MI
to make
MIU
didn't destroy
MI
But in Typogenetics, as in real genetics, the scheme is quite a bit trickier. We do begin with some arbitrary strand, somewhat like an axiom in a formal system. But we have, initially, no "rules of inference"-that is, no enzymes. However, we can
translate
each strand into one or more enzymes! Thus, the strands themselves will dictate the operations which will be performed upon them, and those operations will in turn produce
new strands which will dictate further enzymes, etc. etc.! This is mixing levels with a vengeance! Think, for the sake of comparison, how different the
MU
-puzzle would have been if each new
theorem
produced could have been turned into a new
rule of inference
by means of some code.
How is this "translation" done? It involves a
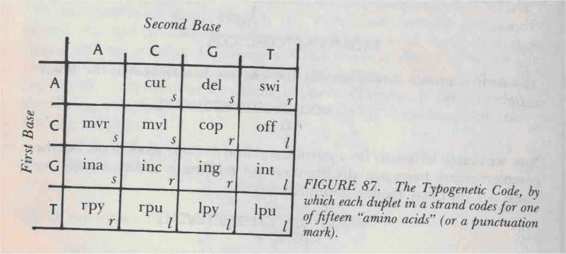
Typogenetic Code
by which adjacent pairs of bases-called "duplets"-in a single strand represent different amino acids. There are sixteen possible duplets:
AA, AC, AG, AT, CA, CC
, etc. And there are fifteen amino acids. The Typogenetic Code is shown in Figure 87.
According to the table, the translation of the duplet GC is "inc" ("insert a C"); that of AT is
"swi" ("switch strands"); and so on. Therefore it becomes clear that a strand can dictate an enzyme very straightforwardly. For example, the strand
TAGATCCAGTCCACATCGA
breaks up into duplets as follows:
TA GA TC CA GT CC AC AT CG A
with the
A
left over at the end. Its translation into an enzyme is: rpy - ina - rpu - mvr - int - mvl - cut - swi - cop.
(Note that the leftover A contributes nothing.)
Tertiary Structure of Enzymes
What about the little letters 's', 'l', and 'r' in the lower righthand corner of each box% They are crucial in determining the enzyme's binding-preference, and in a peculiar way. In order to figure out what letter an enzyme likes to bind to, you have to figure out the enzyme's "tertiary structure", which is itself determined by the enzyme's "primary structure". By its
primary structure
is meant its amino acid sequence. By its tertiary structure is meant the way it likes to "fold up". The point is that enzymes don't like being in straight lines, as we have so far exhibited them. At each internal amino acid (all but the two ends), there is a possibility of a "kink", which is dictated by the letters in the corners. In particular, '1' and 'r' stand for "left"
and "right", and 's' stands for "straight". So let us take our most recent sample enzyme, and let it fold itself up to show its tertiary structure. We will start with the enzyme's primary structure, and move along it from left to right. At each amino acid whose corner-letter is '1'
we'll put a left turn, for those with 'r', we'll put a right turn, and at 's' we'll put no turn. In Figure 88 is shown the two-dimensional conformation for our enzyme.
Cop
swi <== cut <== mvl <== int
mvr
rpy -==> ima ==> rpu
FIGURE 88. The tertiary structure of a typoenzyme.
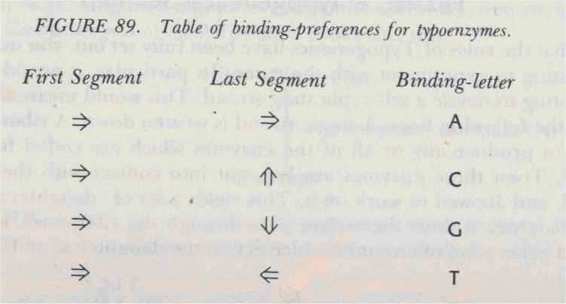
Note the left-kink at "rpu", the right-kink at "swi", and so on. Notice also that the first segment ("rpy z> ina") and the last segment ("swi => cop") are perpendicular. This is the key to the binding-preference. In fact, the
relative orientation of the first and last segments
of an enzyme's tertiary structure determines the binding-preference of the enzyme. We can always orient the enzyme so that its first segment points to the right. If we do so, then the last segment determines the binding-preference, as shown in Figure 89.
So in our case, we have an enzyme which likes the letter C. If, in folding up, an enzyme happens to cross itself, that's okay-just think of it as going under or over itself. Notice that all its amino acids play a role in the determination of an enzyme's tertiary structure.
Punctuation, Genes, and Ribosomes
Now one thing remains to he explained. Why is there a blank in box
AA
of the typogenetic Code' The answer is that the duplet
AA
acts as a punctuation mark inside a strand, and it signals the end of the code for an enzyme.
That is to say, one strand may code for two or more enzymes if it has one or more duplets
AA
in it. For example, the strand
CG GA TA CT AA AC CG A
Codes for two enzymes
cop - ina - rpy - off
and
cut – cop
with the
AA
serving to divide the strand up into two "genes". The definition of gene is: that portion of a strand which codes for a single enzyme. Note that the mere presence of
AA
inside a strand does not mean that the strand codes for two enzymes. For instance,
CAAG
codes for "mvr - del". The
AA
begins on an even-numbered unit and therefore is not read as a duplet!
The mechanism which reads strands and produces the enzymes which are coded inside them is called a ribosome. (In Typogenetics, the player of the game does the work of'
the ribosomes.) Ribosomes are not in any way responsible for the
tertiary
structure of enzymes, for that is entirely determined once the
primary
structure is created. Incidentally, the process of
translation
always goes
from strands to enzymes
, and never in the reverse direction.
Puzzle: A Typogenetical Self-Rep
Now that the rules of Typogenetics have been fully set out, you may find it interesting to experiment with the game. In particular, it would he most interesting to devise a self-replicating strand. This would mean something along the following lines. A single strand is written down. A ribosome acts on it, to produce any or all of the enzymes which are coded for in the strand. Then those enzymes are brought into contact with the original strand, and allowed to work on it. This yields a set of "daughter strands". The daughter strands themselves pass through the rihosomes, to yield a second generation of enzymes, which act on the daughter strands; and the

